bayesNMF: Gibbs sampling for Bayesian Nonnegative Matrix Factorization with automatically learned rank and optional Metropolis-Hastings updates for computational efficiency.
Introduced in the paper "bayesNMF: Fast Bayesian Poisson NMF with Automatically Learned Rank Applied to Mutational Signatures".
For code to reproduce all results in the paper, see the bayesNMF_PAPER respository.
library(devtools)
devtools::install_github("jennalandy/bayesNMF")
library(bayesNMF)
Please see our tutorial
Model details and advanced options can be found here.
The bayesNMF R software package implements an automated convergence detection algorithm and reports convergence metrics in a log file and with trace plots (Figure Panel A). This makes the software simple to use, reduces subjectivity of sampler stopping, and ensures all models reach a similar degree of convergence so results can be compared fairly. Full details of model specifications, Gibbs updates, and hyperparameters are available in paper Appendix A. Details of convergence, inference, and reference assignment are in paper Appendix B.
The bayesNMF R software package allows users to fit all models defined in this paper with the bayesNMF function. Model specifications can be adjusted by the likelihood, prior, and MH parameters. The rank can be a fixed value or a range vector, in which case rank_method specifies whether minBIC, SBFI, or BFI is used to learn rank. Users are also able to set hyperprior parameters or specify initial values of parameters and prior parameters.
The bayesNMF R package allows users to visualize results and optionally compare them to a set of reference signatures (the default are COSMIC v3.3.1 SBS signatures). Figure Panel B provides an example output of the reference assignment.
The plot function creates multiple visualizations comparing the sampler results to the reference. Figure Panel C shows a cosine similarity heatmap between estimates and assigned reference signatures, a dot plot highlighting the median number of mutations attributed to each signature and posterior mean cosine similarity, as well as estimated mutational signatures with posterior uncertainty. Additional functions, plot variations, and a visual diagnostic for label switching are documented in the package vignettes and in Paper Appendix B.5.
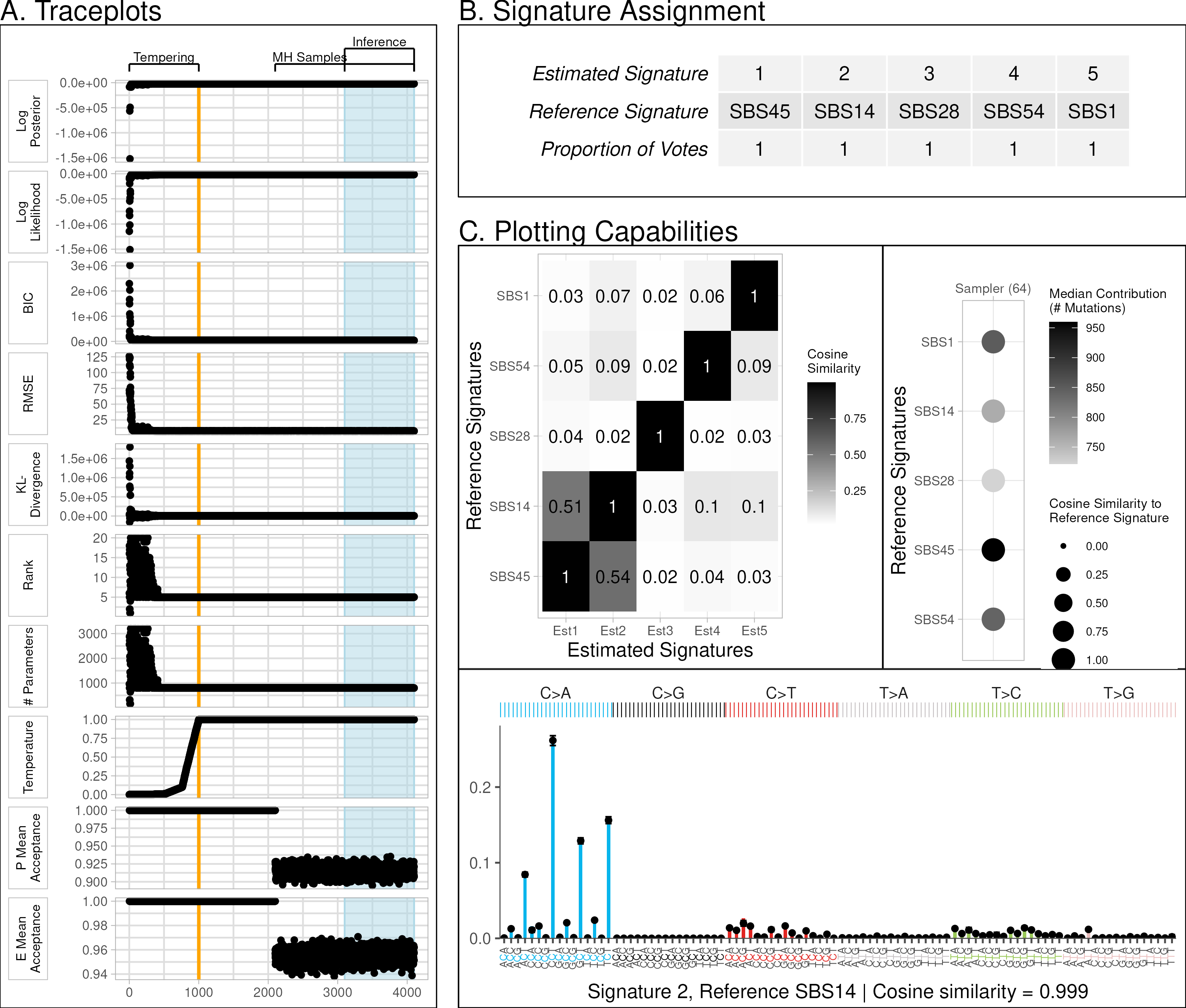 Illustration of software package capabilities using
Illustration of software package capabilities using bayesNMF Poisson-Truncated Normal+MH SBFI on simulated data. A. Posterior diagnostic traceplots. B. Reference assignment using posterior ensemble with majority voting. C. Visualization suite, including similarity heatmaps, contribution summaries, and reconstructed signatures (bar chart of aligned reference, points for final estimates, and error bars for 95% credible intervals).